Juho Ylä-Jääski

Links
Identification of Gene Regulatory Network
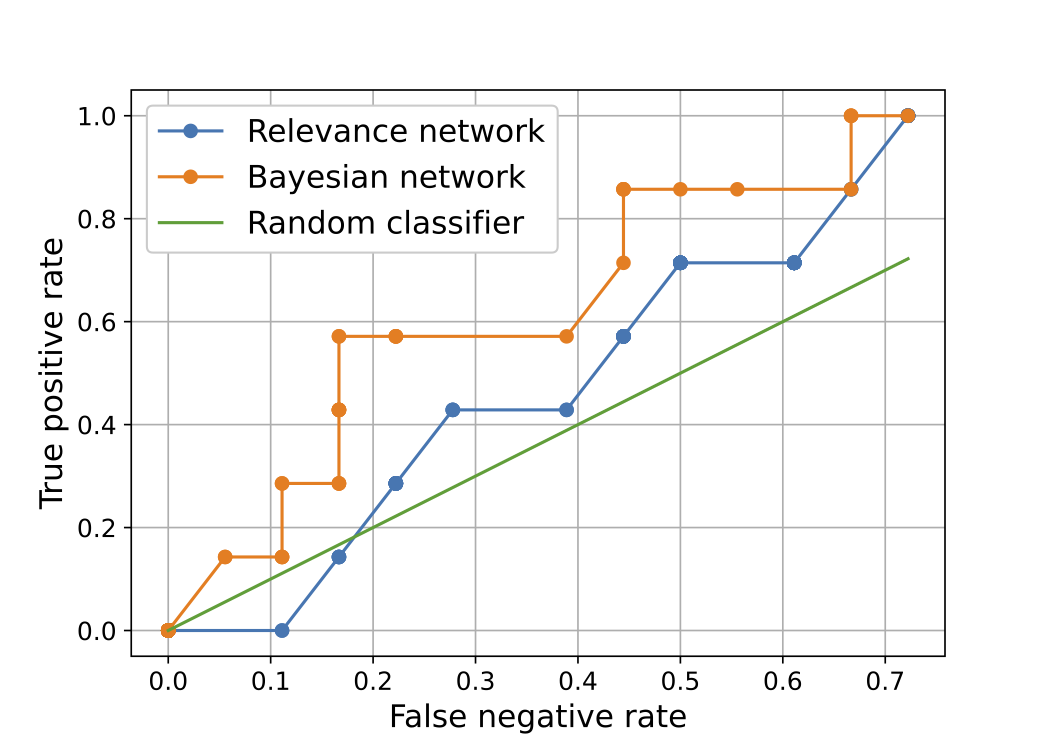
Project description: This project was a part of the course CS-E5885 - Modeling Biological Networks D. The goal of the project was to study computational/statistical methods which were then used to infer the structure of a 5-gene network using gene expression time-course data only. The data was provided in the course. Our first idea was to tackle the problem with two simple approaches - relevance network and Bayesian network. With these simple approaches only slightly better results in comparison to a random classifier were achieved, thus, we also did a more sophisticated approach with an ODE system.
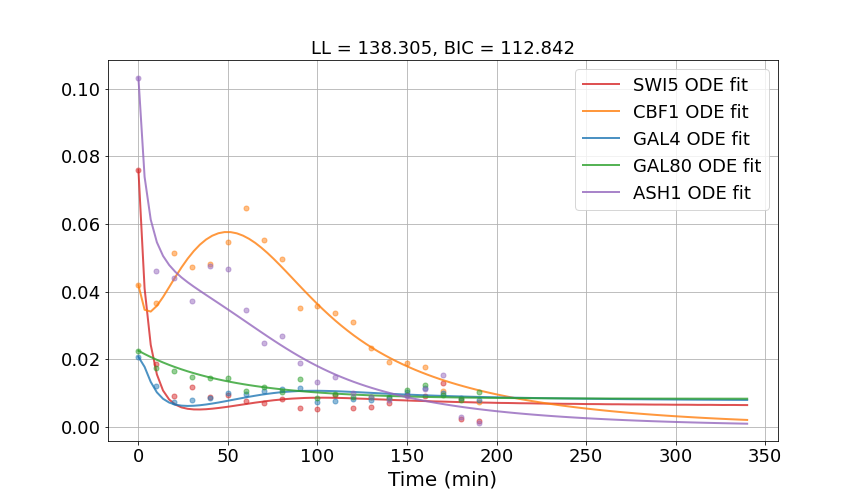
Results: The solved ODEs with optimized parameters fit the time-series data reasonably well and capture the overall dynamics. The declines in all gene expressions are present in the fitted functions and the final values around time 190 min match closely with the data. Even some interesting dips and peaks in the data are captured in the functions of GAL4 and CBF1. It is, however, possible that the fitted functions are smoother than the phenomena in reality. One possible limitation of our approach is the mass-action kinetics which does not model the details of biochemical reactions